Machine Learning for Respiratory Health Monitoring on Edge Devices
Project Overview
This report details a multi-semester senior design project focused on creating a device for real-time respiratory health analysis using machine learning on edge devices. The project involved a team of four computer science students under the guidance of Dr. Zhewei Wang (Machine Learning) and Dr. Savas Kaya (Electrical Engineering). My primary responsibilities included data preprocessing, model creation and optimization, and hardware deployment.
The project progressed through six distinct phases, beginning with foundational research and culminating in a final, integrated prototype on the Google Coral Dev Board.
Phase 1: Research, and Speech Commands Model Creation/Compression
(October 21 - November 18, 2024)
The project began with an accelerated course in machine learning principles, covering neural networks, backpropagation, and CNN/RNN architectures.
Data Preprocessing and Feature Extraction
The first practical step was to establish a data pipeline for a proof-of-concept model using the “Mini Speech Commands” dataset. I developed a Python script (waveform_transformations.py) to process raw audio into various 2D representations suitable for a CNN, including standard spectrograms, Mel Spectrograms, and Mel-Frequency Cepstral Coefficients (MFCCs).
Initial CNN Model Development
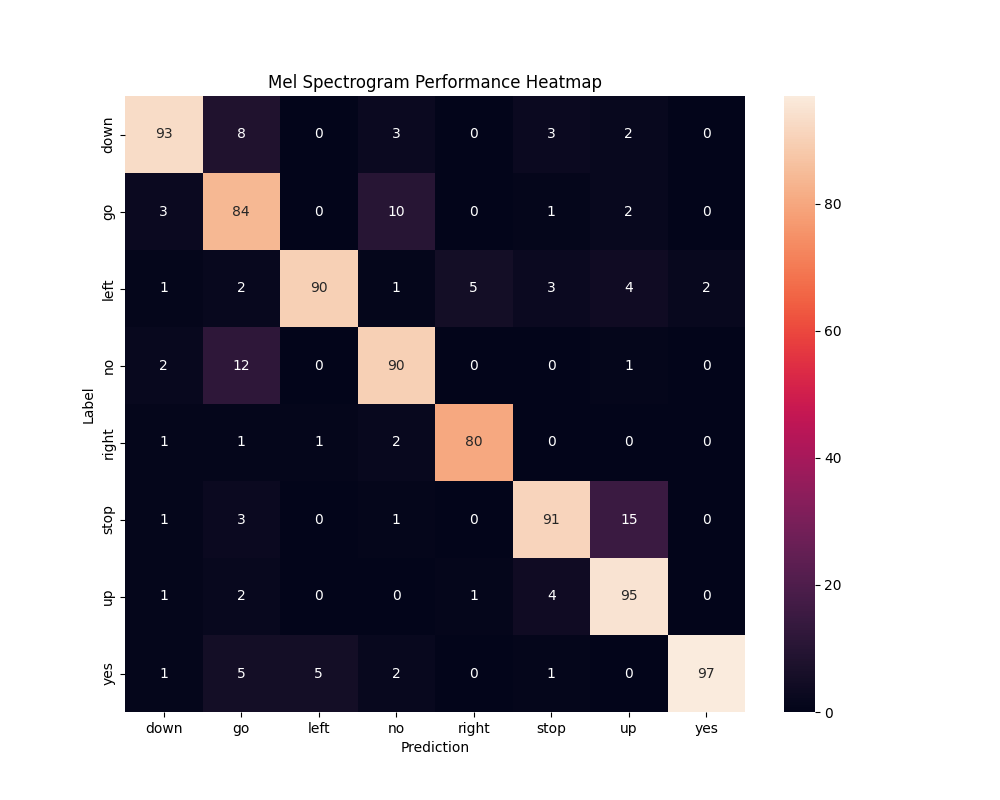
Using TensorFlow/Keras, I built and trained a CNN (TF_CNN.py) to classify the 8 keywords. After testing the different feature types, I determined that Mel Spectrograms yielded the highest performance, achieving approximately 90% validation accuracy and confirming the viability of our approach.
Hardware Research
Concurrently, I researched hardware and selected the Raspberry Pi 5 as a powerful single-board computer (SBC) and the Raspberry Pi Pico as a low-power microcontroller target.
Model Optimization and Challenges
To prepare for deployment, I successfully applied post-training quantization (quantize.py), shrinking the model from 18.6 MB to 1.55 MB with minimal accuracy loss. An attempt to deploy to a Banana Pi M4 was abandoned due to significant dependency and support issues, a key lesson in hardware selection.
Live Inference Demo
This phase concluded with a real-time demo (quantized_CNN_demo.py) that captured microphone audio on a laptop, processed it, and fed it to the quantized TFLite model for live inference, successfully proving the end-to-end concept.
Phase 2: Respiratory Model Creation and Optimization
(November 18, 2024 - December 2, 2024)
This phase shifted to the project’s primary goal: classifying respiratory sounds.
Dataset Acquisition and Initial Model
I sourced the “Respiratory Sound Database” from Kaggle and wrote a script (format_dataset-1.py) to parse annotations and slice the audio into clips containing crackles, wheezes, both, or normal breathing. An initial CNN model (respiratory_CNN-1.py) trained on this data performed poorly, highlighting the need for a more rigorous methodology.
Systematic Optimization
To improve performance, I rewrote the training script (respiratory_CNN-2.py) to systematically test every combination of dataset variants (variable vs. 8-second padded clips), preprocessing methods (Spectrogram, Mel Spectrogram, MFCC), and seven different TensorFlow optimizers. This exhaustive search allowed me to empirically identify the optimal configuration: the Adam optimizer with Mel Spectrograms.
Advanced Model Iteration
To meet a new accuracy target of >70%, I further refined the process. A new script (format_dataset-3.py) filtered the dataset to only include high-quality samples (44.1kHz). I then improved the model architecture (respiratory_CNN-3.py) with LeakyReLU activations and SpatialDropout2D layers. These changes successfully pushed the model’s accuracy above the required threshold, confirmed with a final evaluation script (CNN_Evaluations-3.py). 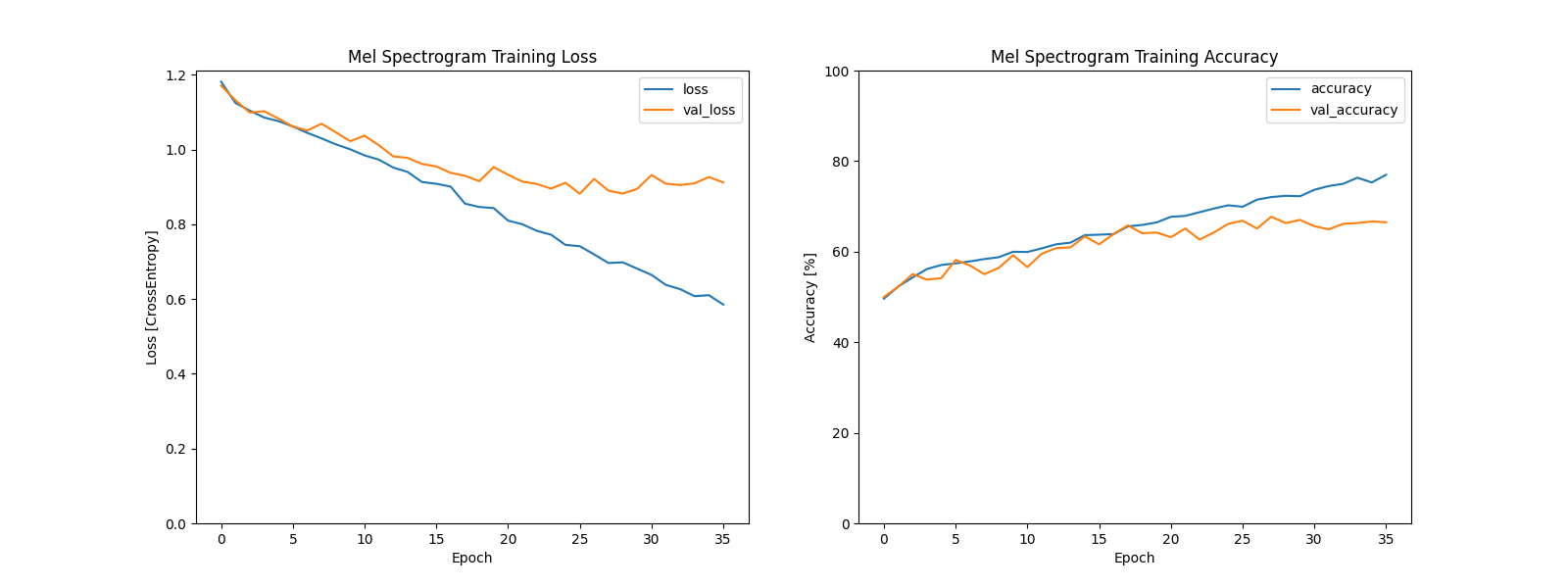
Phase 3: Pico Deployment and Memory Constraints
(January 20, 2025 - February 3, 2025)
The first hardware target was the resource-constrained Raspberry Pi Pico.
Pivoting to a Simpler Model
I first attempted to deploy a stripped-down, quantized version of the respiratory model (respiratory_CNN-4.py). While the model could be stored in the Pico’s flash memory, there was insufficient RAM (264KB) to load the interpreter and run inference. This forced a strategic pivot to the much smaller speech commands model to validate the deployment workflow.
PC-to-Pico Inference Pipeline
I engineered a system where a PC handled audio capture and streaming. A Python script (send_data.py) transmitted audio data over UART to the Pico. On the device, an Arduino script (sketch_feb4a.ino) loaded the TFLite model, received the data, ran inference using the TensorFlow Lite for Microcontrollers interpreter, and sent the prediction back to the PC. This successfully demonstrated a complete inference pipeline on a microcontroller, providing valuable lessons on the trade-offs between model complexity and hardware limitations.
Phase 4: Transition to 1D CNN Architecture
(February 3, 2025 - February 24, 2025)
To address the memory issues and improve efficiency, we transitioned from 2D CNNs (processing images) to 1D CNNs (processing raw audio waveforms), eliminating the need for on-device spectrogram generation.
Model Development and Enhancement
I first adapted a baseline 1D model for respiratory data (respiratory_CNN_1D-1.py). When its accuracy proved insufficient, I engineered an improved version (respiratory_CNN_1D-2.py) that integrated a Multi-Head Temporal Attention mechanism. This allowed the model to focus on the most salient parts of the audio waveform, significantly improving classification accuracy.
Multi-Head Temporal Attention (respiratory_CNN_1D-2.py)
Data Augmentation Pipeline
To further enhance robustness, I implemented a full data augmentation pipeline in PyTorch (Respiratory_CNN_1D-3.py). During training, this script would randomly apply transformations to the audio samples, including adding Gaussian noise, applying random gain, time-shifting, and inverting polarity. This process created a more resilient model, marking a successful pivot to a more efficient 1D architecture.
Data Augmentation Pipeline (respiratory_CNN_1D-3.py)
Phase 5: ESP32 Prototyping and System Integration
(February 24, 2025 - March 17, 2025)
The next prototype was built on the more powerful Keyestudio ESP32 Inventor Starter Kit.
Platform Setup and Integration
I flashed the ESP32 with MicroPython and the TensorFlow Lite interpreter. I then developed a series of scripts to test and integrate each peripheral: microphone input (mic_test.py), LCD output (lcd_test.py), and WiFi communication to a Flask server (wifi_test.py, server.py).
End-to-End System Demo
Finally, I combined these modules into a single script (1D_CNN_ESP32.py). This program orchestrated a complete workflow: on a button press, it recorded 1 second of audio, ran inference with the speech commands model, displayed the prediction on the LCD, and sent the result to a server over WiFi. This created a fully functional, self-contained embedded ML prototype that served as the blueprint for the final device.
Phase 6: Final Deployment on Google Coral
(March 17 - April 14, 2025)
The final phase utilized the Google Coral Dev Board, leveraging its Edge TPU for accelerated inference.
Hardware and Model Pivots
I began by deploying TFLite versions of our respiratory, PPG, and ECG models to the Coral. For data acquisition, I 3D printed and assembled an open-source “Stethogram,” a digital stethoscope. While it could detect a heartbeat, it was unable to reliably capture lung sounds.
Simultaneously, I discovered a flaw in our respiratory model’s training data (1-second clips instead of 8) and found its on-device performance lacking. This prompted a final pivot to a high-performing public model on Kaggle that classified respiratory diseases.
Overcoming Compatibility Issues
This new model presented a major challenge: it was built with a version of TensorFlow incompatible with the Coral’s Edge TPU compiler. To solve this, I set up a legacy virtual environment with older versions of Python and TensorFlow (2.5). Within this environment, I re-trained the model (new_resp_model.py), quantized it, and successfully compiled it for the Edge TPU (quantize_tf2-5.py).
Final Integrated System
The final script (resp_disease_model.py) brought everything together. It captures audio from the microphone, applies a Butterworth filter, and feeds the data to the Edge TPU-accelerated disease model for real-time inference. The program then prints the predicted probabilities for six different respiratory diseases. This fully self-contained system marked the successful conclusion of the project.
Final Inference Loop (resp_disease_model.py)